Datasets:
Tasks:
Image Segmentation
Modalities:
Image
Formats:
parquet
Sub-tasks:
instance-segmentation
Languages:
English
Size:
1K - 10K
ArXiv:
License:
File size: 4,492 Bytes
b9a02ac 5389c54 b9a02ac |
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 |
---
dataset_info:
features:
- name: image
dtype:
image:
mode: RGB
- name: instances
sequence:
image:
mode: '1'
- name: categories
sequence:
class_label:
names:
'0': Neoplastic
'1': Inflammatory
'2': Connective
'3': Dead
'4': Epithelial
- name: tissue
dtype:
class_label:
names:
'0': Adrenal Gland
'1': Bile Duct
'2': Bladder
'3': Breast
'4': Cervix
'5': Colon
'6': Esophagus
'7': Head & Neck
'8': Kidney
'9': Liver
'10': Lung
'11': Ovarian
'12': Pancreatic
'13': Prostate
'14': Skin
'15': Stomach
'16': Testis
'17': Thyroid
'18': Uterus
splits:
- name: fold1
num_bytes: 283673837.64
num_examples: 2656
- name: fold2
num_bytes: 267595457.439
num_examples: 2523
- name: fold3
num_bytes: 293079722.82
num_examples: 2722
download_size: 1665092597
dataset_size: 844349017.8989999
configs:
- config_name: default
data_files:
- split: fold1
path: data/fold1-*
- split: fold2
path: data/fold2-*
- split: fold3
path: data/fold3-*
license: cc-by-nc-sa-4.0
task_categories:
- image-segmentation
task_ids:
- instance-segmentation
language:
- en
tags:
- medical
- cell nuclei
- H&E
pretty_name: PanNuke
size_categories:
- 1K<n<10K
paperswithcode_id: pannuke
---
# PanNuke
[](https://warwick.ac.uk/fac/cross_fac/tia/data/pannuke)
## Dataset Description
- **Homepage:** [PanNuke Dataset for Nuclei Instance Segmentation and Classification](https://warwick.ac.uk/fac/cross_fac/tia/data/pannuke)
- **Leaderboard:** [Panoptic Segmentation](https://paperswithcode.com/sota/panoptic-segmentation-on-pannuke)
## Description
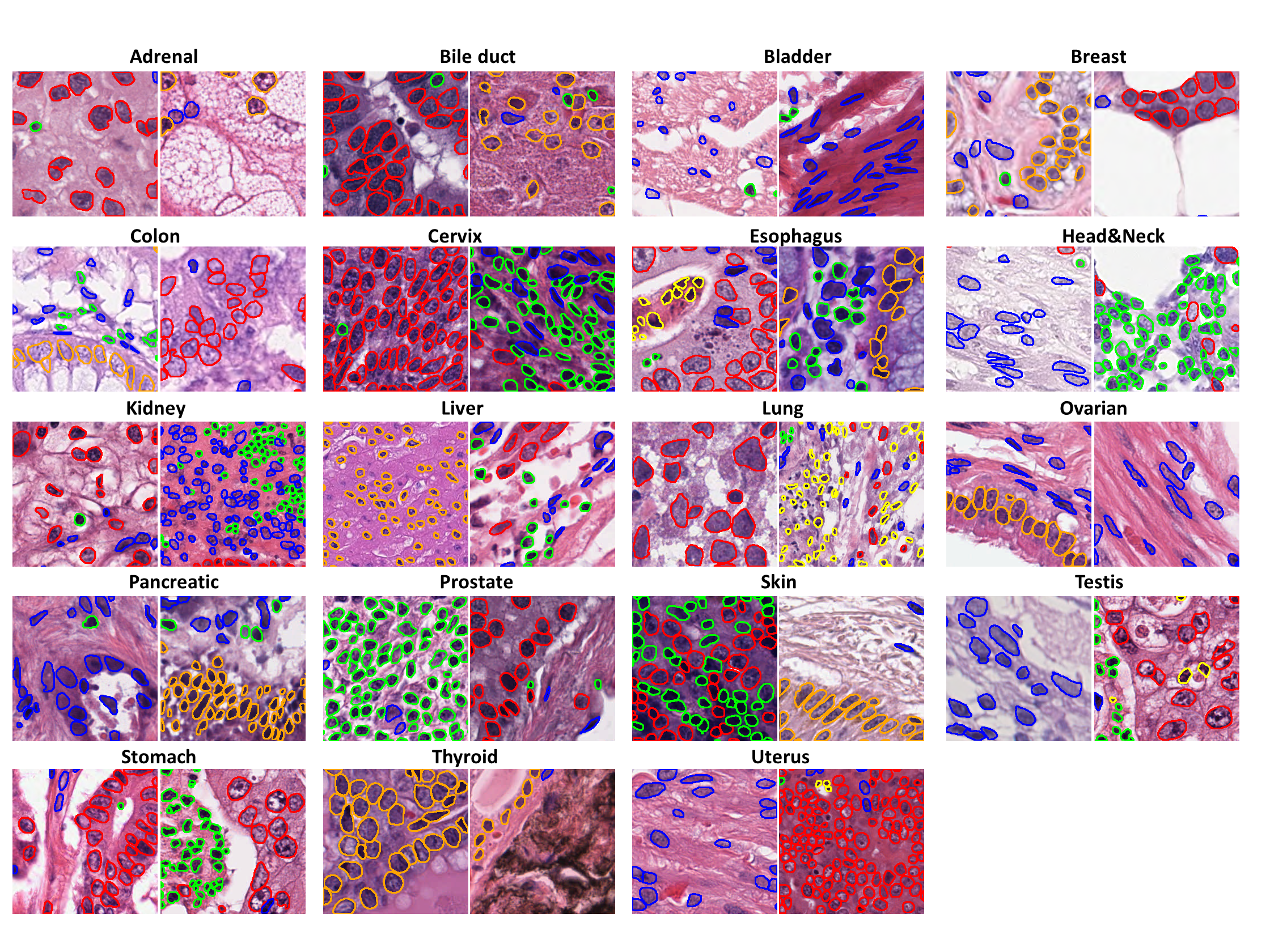
PanNuke is a semi-automatically generated dataset for nuclei instance segmentation and classification, providing comprehensive nuclei annotations across 19 tissue types and 5 distinct cell categories. The dataset includes a total of **189,744 labeled nuclei**, each accompanied by an instance segmentation mask, and contains **7,901 images**, each sized **256×256 pixels**. The images were captured at **x40 magnification** with a resolution of **0.25 µm/pixel**. The dataset is highly imbalanced, with the **"Dead" nuclei category** being particularly underrepresented.
Please note that the dataset was created by extracting patches from whole-slide images (WSIs). As a result, some nuclei located at the edges of patches may be cropped, with fewer than 10 visible pixels in certain cases.
## Dataset Structure
The dataset is organized into three folds: `fold1`, `fold2`, and `fold3`, consistent with the original dataset structure. Each fold contains data in a tabular format with the following four columns:
- **`image`**: The RGB tile of the sample.
- **`instances`**: A list of nuclei instances. Each instance represents exactly one nucleus and is in binary format (`1` - nucleus, `0` - background)
- **`categories`**: An integer class label for each nucleus, corresponding to one of the following categories:
0. Neoplastic
1. Inflammatory
2. Connective
3. Dead
4. Epithelial
- **`tissue`**: The integer tissue type from which the sample originates, belonging to one of these categories:
0. Adrenal Gland
1. Bile Duct
2. Bladder
3. Breast
4. Cervix
5. Colon
6. Esophagus
7. Head & Neck
8. Kidney
9. Liver
10. Lung
11. Ovarian
12. Pancreatic
13. Prostate
14. Skin
15. Stomach
16. Testis
17. Thyroid
18. Uterus
## Citation
```bibtex
@inproceedings{gamper2019pannuke,
title={PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification},
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Benes, Ksenija and Khuram, Ali and Rajpoot, Nasir},
booktitle={European Congress on Digital Pathology},
pages={11--19},
year={2019},
organization={Springer}
}
```
```bibtex
@article{gamper2020pannuke,
title={PanNuke Dataset Extension, Insights and Baselines},
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Graham, Simon and Jahanifar, Mostafa and Khurram, Syed Ali and Azam, Ayesha and Hewitt, Katherine and Rajpoot, Nasir},
journal={arXiv preprint arXiv:2003.10778},
year={2020}
}
``` |