Datasets:
image
imagewidth (px) 256
256
| instances
images listlengths 0
276
| categories
sequencelengths 0
276
| tissue
class label 19
classes |
|---|---|---|---|
[
0,
0,
0,
0,
1,
1,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
1,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
0,
0,
0,
1,
1,
2,
2
] | 3Breast
|
||
[
0,
0,
1,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
1,
1,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
2,
2
] | 3Breast
|
||
[
0,
0,
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
1,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
1,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
1,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
1,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
1,
2,
2
] | 3Breast
|
||
[
0,
0,
1,
1,
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
1,
1,
1,
1,
1,
1,
1,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
1,
1,
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
2
] | 3Breast
|
||
[
0,
0,
1,
1,
1,
1,
1,
1,
1,
1,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
1,
1,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
1,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
4
] | 3Breast
|
||
[
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0
] | 3Breast
|
||
[
0,
0,
0,
0,
1,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
0,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
0,
0,
0,
0,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
1,
1,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2
] | 3Breast
|
||
[
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
1,
1,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
1,
2,
2,
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
2,
2,
2,
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
2,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
||
[
2,
2,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4,
4
] | 3Breast
|
PanNuke
Description
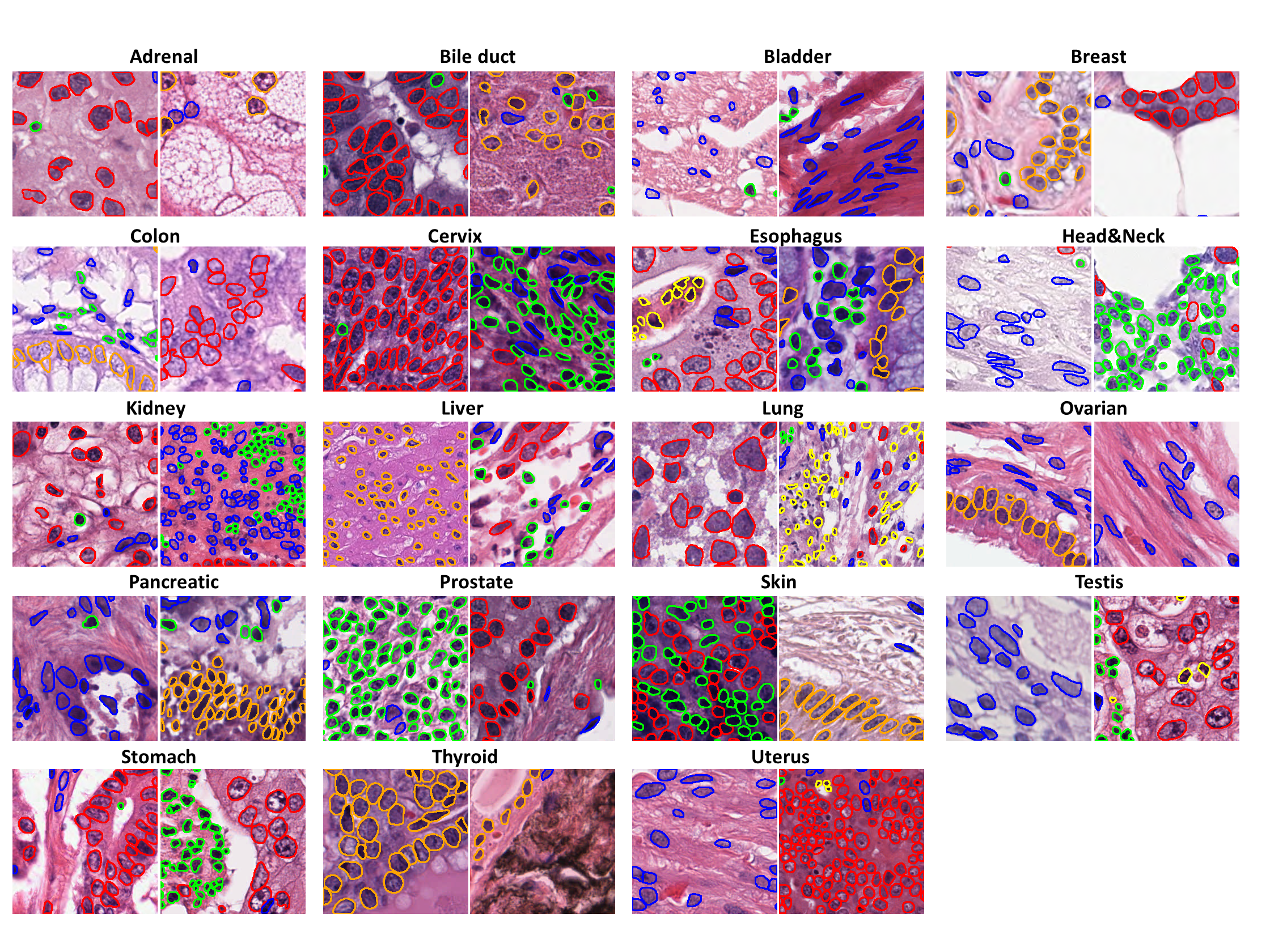
PanNuke is a semi-automatically generated dataset for nuclei instance segmentation and classification, providing comprehensive nuclei annotations across 19 tissue types and 5 distinct cell categories. The dataset includes a total of 189,744 labeled nuclei, each accompanied by an instance segmentation mask, and contains 7,901 images, each sized 256×256 pixels. The images were captured at x40 magnification with a resolution of 0.25 µm/pixel. The dataset is highly imbalanced, with the "Dead" nuclei category being particularly underrepresented.
Please note that the dataset was created by extracting patches from whole-slide images (WSIs). As a result, some nuclei located at the edges of patches may be cropped, with fewer than 10 visible pixels in certain cases.
Dataset Structure
The dataset is organized into three folds: fold1, fold2, and fold3, consistent with the original dataset structure. Each fold contains data in a tabular format with the following four columns:
image: The RGB tile of the sample.instances: A list of nuclei instances. Each instance represents exactly one nucleus and is in binary format (1- nucleus,0- background)categories: An integer class label for each nucleus, corresponding to one of the following categories:- Neoplastic
- Inflammatory
- Connective
- Dead
- Epithelial
tissue: The integer tissue type from which the sample originates, belonging to one of these categories:- Adrenal Gland
- Bile Duct
- Bladder
- Breast
- Cervix
- Colon
- Esophagus
- Head & Neck
- Kidney
- Liver
- Lung
- Ovarian
- Pancreatic
- Prostate
- Skin
- Stomach
- Testis
- Thyroid
- Uterus
Citation
@inproceedings{gamper2019pannuke,
title={PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification},
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Benes, Ksenija and Khuram, Ali and Rajpoot, Nasir},
booktitle={European Congress on Digital Pathology},
pages={11--19},
year={2019},
organization={Springer}
}
@article{gamper2020pannuke,
title={PanNuke Dataset Extension, Insights and Baselines},
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Graham, Simon and Jahanifar, Mostafa and Khurram, Syed Ali and Azam, Ayesha and Hewitt, Katherine and Rajpoot, Nasir},
journal={arXiv preprint arXiv:2003.10778},
year={2020}
}
- Downloads last month
- 47