Datasets:
Tasks:
Image Segmentation
Modalities:
Image
Formats:
parquet
Sub-tasks:
instance-segmentation
Languages:
English
Size:
1K - 10K
ArXiv:
License:
Commit
·
b9a02ac
verified
·
0
Parent(s):
Initial commit
Browse files- .gitattributes +59 -0
- README.md +148 -0
- data/fold1-00000-of-00001.parquet +3 -0
- data/fold2-00000-of-00001.parquet +3 -0
- data/fold3-00000-of-00001.parquet +3 -0
- gen_script.py +127 -0
.gitattributes
ADDED
|
@@ -0,0 +1,59 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
*.7z filter=lfs diff=lfs merge=lfs -text
|
| 2 |
+
*.arrow filter=lfs diff=lfs merge=lfs -text
|
| 3 |
+
*.bin filter=lfs diff=lfs merge=lfs -text
|
| 4 |
+
*.bz2 filter=lfs diff=lfs merge=lfs -text
|
| 5 |
+
*.ckpt filter=lfs diff=lfs merge=lfs -text
|
| 6 |
+
*.ftz filter=lfs diff=lfs merge=lfs -text
|
| 7 |
+
*.gz filter=lfs diff=lfs merge=lfs -text
|
| 8 |
+
*.h5 filter=lfs diff=lfs merge=lfs -text
|
| 9 |
+
*.joblib filter=lfs diff=lfs merge=lfs -text
|
| 10 |
+
*.lfs.* filter=lfs diff=lfs merge=lfs -text
|
| 11 |
+
*.lz4 filter=lfs diff=lfs merge=lfs -text
|
| 12 |
+
*.mds filter=lfs diff=lfs merge=lfs -text
|
| 13 |
+
*.mlmodel filter=lfs diff=lfs merge=lfs -text
|
| 14 |
+
*.model filter=lfs diff=lfs merge=lfs -text
|
| 15 |
+
*.msgpack filter=lfs diff=lfs merge=lfs -text
|
| 16 |
+
*.npy filter=lfs diff=lfs merge=lfs -text
|
| 17 |
+
*.npz filter=lfs diff=lfs merge=lfs -text
|
| 18 |
+
*.onnx filter=lfs diff=lfs merge=lfs -text
|
| 19 |
+
*.ot filter=lfs diff=lfs merge=lfs -text
|
| 20 |
+
*.parquet filter=lfs diff=lfs merge=lfs -text
|
| 21 |
+
*.pb filter=lfs diff=lfs merge=lfs -text
|
| 22 |
+
*.pickle filter=lfs diff=lfs merge=lfs -text
|
| 23 |
+
*.pkl filter=lfs diff=lfs merge=lfs -text
|
| 24 |
+
*.pt filter=lfs diff=lfs merge=lfs -text
|
| 25 |
+
*.pth filter=lfs diff=lfs merge=lfs -text
|
| 26 |
+
*.rar filter=lfs diff=lfs merge=lfs -text
|
| 27 |
+
*.safetensors filter=lfs diff=lfs merge=lfs -text
|
| 28 |
+
saved_model/**/* filter=lfs diff=lfs merge=lfs -text
|
| 29 |
+
*.tar.* filter=lfs diff=lfs merge=lfs -text
|
| 30 |
+
*.tar filter=lfs diff=lfs merge=lfs -text
|
| 31 |
+
*.tflite filter=lfs diff=lfs merge=lfs -text
|
| 32 |
+
*.tgz filter=lfs diff=lfs merge=lfs -text
|
| 33 |
+
*.wasm filter=lfs diff=lfs merge=lfs -text
|
| 34 |
+
*.xz filter=lfs diff=lfs merge=lfs -text
|
| 35 |
+
*.zip filter=lfs diff=lfs merge=lfs -text
|
| 36 |
+
*.zst filter=lfs diff=lfs merge=lfs -text
|
| 37 |
+
*tfevents* filter=lfs diff=lfs merge=lfs -text
|
| 38 |
+
# Audio files - uncompressed
|
| 39 |
+
*.pcm filter=lfs diff=lfs merge=lfs -text
|
| 40 |
+
*.sam filter=lfs diff=lfs merge=lfs -text
|
| 41 |
+
*.raw filter=lfs diff=lfs merge=lfs -text
|
| 42 |
+
# Audio files - compressed
|
| 43 |
+
*.aac filter=lfs diff=lfs merge=lfs -text
|
| 44 |
+
*.flac filter=lfs diff=lfs merge=lfs -text
|
| 45 |
+
*.mp3 filter=lfs diff=lfs merge=lfs -text
|
| 46 |
+
*.ogg filter=lfs diff=lfs merge=lfs -text
|
| 47 |
+
*.wav filter=lfs diff=lfs merge=lfs -text
|
| 48 |
+
# Image files - uncompressed
|
| 49 |
+
*.bmp filter=lfs diff=lfs merge=lfs -text
|
| 50 |
+
*.gif filter=lfs diff=lfs merge=lfs -text
|
| 51 |
+
*.png filter=lfs diff=lfs merge=lfs -text
|
| 52 |
+
*.tiff filter=lfs diff=lfs merge=lfs -text
|
| 53 |
+
# Image files - compressed
|
| 54 |
+
*.jpg filter=lfs diff=lfs merge=lfs -text
|
| 55 |
+
*.jpeg filter=lfs diff=lfs merge=lfs -text
|
| 56 |
+
*.webp filter=lfs diff=lfs merge=lfs -text
|
| 57 |
+
# Video files - compressed
|
| 58 |
+
*.mp4 filter=lfs diff=lfs merge=lfs -text
|
| 59 |
+
*.webm filter=lfs diff=lfs merge=lfs -text
|
README.md
ADDED
|
@@ -0,0 +1,148 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
---
|
| 2 |
+
dataset_info:
|
| 3 |
+
features:
|
| 4 |
+
- name: image
|
| 5 |
+
dtype:
|
| 6 |
+
image:
|
| 7 |
+
mode: RGB
|
| 8 |
+
- name: instances
|
| 9 |
+
sequence:
|
| 10 |
+
image:
|
| 11 |
+
mode: '1'
|
| 12 |
+
- name: categories
|
| 13 |
+
sequence:
|
| 14 |
+
class_label:
|
| 15 |
+
names:
|
| 16 |
+
'0': Neoplastic
|
| 17 |
+
'1': Inflammatory
|
| 18 |
+
'2': Connective
|
| 19 |
+
'3': Dead
|
| 20 |
+
'4': Epithelial
|
| 21 |
+
- name: tissue
|
| 22 |
+
dtype:
|
| 23 |
+
class_label:
|
| 24 |
+
names:
|
| 25 |
+
'0': Adrenal Gland
|
| 26 |
+
'1': Bile Duct
|
| 27 |
+
'2': Bladder
|
| 28 |
+
'3': Breast
|
| 29 |
+
'4': Cervix
|
| 30 |
+
'5': Colon
|
| 31 |
+
'6': Esophagus
|
| 32 |
+
'7': Head & Neck
|
| 33 |
+
'8': Kidney
|
| 34 |
+
'9': Liver
|
| 35 |
+
'10': Lung
|
| 36 |
+
'11': Ovarian
|
| 37 |
+
'12': Pancreatic
|
| 38 |
+
'13': Prostate
|
| 39 |
+
'14': Skin
|
| 40 |
+
'15': Stomach
|
| 41 |
+
'16': Testis
|
| 42 |
+
'17': Thyroid
|
| 43 |
+
'18': Uterus
|
| 44 |
+
splits:
|
| 45 |
+
- name: fold1
|
| 46 |
+
num_bytes: 283673837.64
|
| 47 |
+
num_examples: 2656
|
| 48 |
+
- name: fold2
|
| 49 |
+
num_bytes: 267595457.439
|
| 50 |
+
num_examples: 2523
|
| 51 |
+
- name: fold3
|
| 52 |
+
num_bytes: 293079722.82
|
| 53 |
+
num_examples: 2722
|
| 54 |
+
download_size: 1665092597
|
| 55 |
+
dataset_size: 844349017.8989999
|
| 56 |
+
configs:
|
| 57 |
+
- config_name: default
|
| 58 |
+
data_files:
|
| 59 |
+
- split: fold1
|
| 60 |
+
path: data/fold1-*
|
| 61 |
+
- split: fold2
|
| 62 |
+
path: data/fold2-*
|
| 63 |
+
- split: fold3
|
| 64 |
+
path: data/fold3-*
|
| 65 |
+
license: cc-by-nc-sa-4.0
|
| 66 |
+
task_categories:
|
| 67 |
+
- image-segmentation
|
| 68 |
+
task_ids:
|
| 69 |
+
- instance-segmentation
|
| 70 |
+
language:
|
| 71 |
+
- en
|
| 72 |
+
tags:
|
| 73 |
+
- medical
|
| 74 |
+
- cell nuclei
|
| 75 |
+
- H&E
|
| 76 |
+
pretty_name: PanNuke
|
| 77 |
+
size_categories:
|
| 78 |
+
- 1K<n<10K
|
| 79 |
+
paperswithcode_id: pannuke
|
| 80 |
+
---
|
| 81 |
+
|
| 82 |
+
# PanNuke
|
| 83 |
+
|
| 84 |
+
[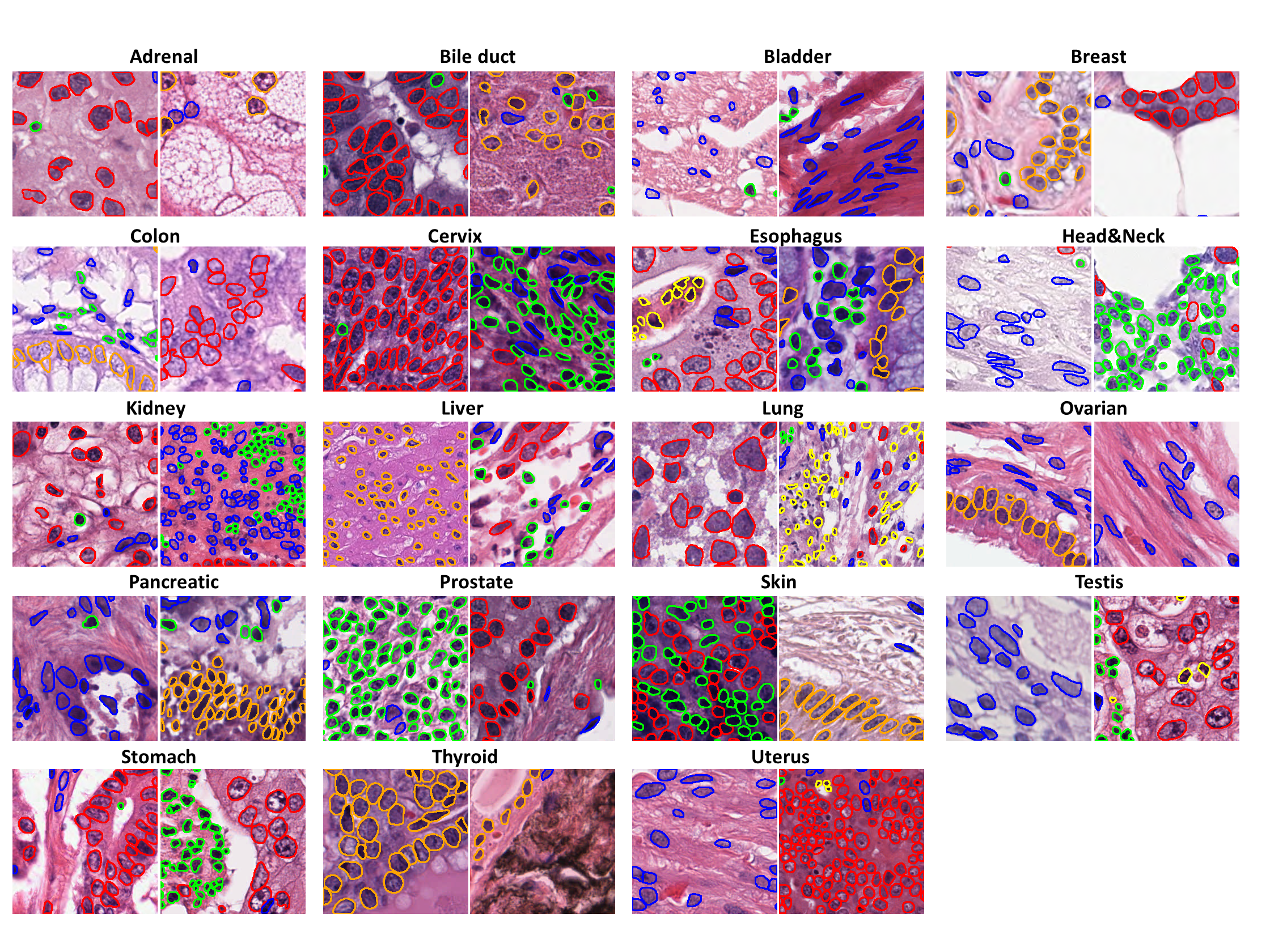](https://warwick.ac.uk/fac/cross_fac/tia/data/pannuke)
|
| 85 |
+
|
| 86 |
+
## Dataset Description
|
| 87 |
+
|
| 88 |
+
- **Homepage:** [PanNuke Dataset for Nuclei Instance Segmentation and Classification](https://warwick.ac.uk/fac/cross_fac/tia/data/pannuke)
|
| 89 |
+
- **Leaderboard:** [Panoptic Segmentation](https://paperswithcode.com/sota/panoptic-segmentation-on-pannuke)
|
| 90 |
+
|
| 91 |
+
## Description
|
| 92 |
+
|
| 93 |
+
PanNuke is a semi automatically generated nuclei instance segmentation and classification dataset with exhaustive nuclei labels across 19 different tissue types. In total the dataset contains 189,744 labeled nuclei, each with an instance segmentation mask.
|
| 94 |
+
|
| 95 |
+
## Dataset Structure
|
| 96 |
+
|
| 97 |
+
The dataset is organized into three folds: `fold1`, `fold2`, and `fold3`, consistent with the original dataset structure. Each fold contains data in a tabular format with the following four columns:
|
| 98 |
+
|
| 99 |
+
- **`image`**: The RGB tile of the sample.
|
| 100 |
+
- **`instances`**: A list of nuclei instances. Each instance represents exactly one nucleus and is in binary format (`1` - nucleus, `0` - background)
|
| 101 |
+
- **`categories`**: An integer class label for each nucleus, corresponding to one of the following categories:
|
| 102 |
+
0. Neoplastic
|
| 103 |
+
1. Inflammatory
|
| 104 |
+
2. Connective
|
| 105 |
+
3. Dead
|
| 106 |
+
4. Epithelial
|
| 107 |
+
- **`tissue`**: The integer tissue type from which the sample originates, belonging to one of these categories:
|
| 108 |
+
0. Adrenal Gland
|
| 109 |
+
1. Bile Duct
|
| 110 |
+
2. Bladder
|
| 111 |
+
3. Breast
|
| 112 |
+
4. Cervix
|
| 113 |
+
5. Colon
|
| 114 |
+
6. Esophagus
|
| 115 |
+
7. Head & Neck
|
| 116 |
+
8. Kidney
|
| 117 |
+
9. Liver
|
| 118 |
+
10. Lung
|
| 119 |
+
11. Ovarian
|
| 120 |
+
12. Pancreatic
|
| 121 |
+
13. Prostate
|
| 122 |
+
14. Skin
|
| 123 |
+
15. Stomach
|
| 124 |
+
16. Testis
|
| 125 |
+
17. Thyroid
|
| 126 |
+
18. Uterus
|
| 127 |
+
|
| 128 |
+
## Citation
|
| 129 |
+
|
| 130 |
+
```bibtex
|
| 131 |
+
@inproceedings{gamper2019pannuke,
|
| 132 |
+
title={PanNuke: an open pan-cancer histology dataset for nuclei instance segmentation and classification},
|
| 133 |
+
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Benes, Ksenija and Khuram, Ali and Rajpoot, Nasir},
|
| 134 |
+
booktitle={European Congress on Digital Pathology},
|
| 135 |
+
pages={11--19},
|
| 136 |
+
year={2019},
|
| 137 |
+
organization={Springer}
|
| 138 |
+
}
|
| 139 |
+
```
|
| 140 |
+
|
| 141 |
+
```bibtex
|
| 142 |
+
@article{gamper2020pannuke,
|
| 143 |
+
title={PanNuke Dataset Extension, Insights and Baselines},
|
| 144 |
+
author={Gamper, Jevgenij and Koohbanani, Navid Alemi and Graham, Simon and Jahanifar, Mostafa and Khurram, Syed Ali and Azam, Ayesha and Hewitt, Katherine and Rajpoot, Nasir},
|
| 145 |
+
journal={arXiv preprint arXiv:2003.10778},
|
| 146 |
+
year={2020}
|
| 147 |
+
}
|
| 148 |
+
```
|
data/fold1-00000-of-00001.parquet
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:84428c1abae5015baf6b324f4927fe8558bbb6610137eb047a335aae7d040f25
|
| 3 |
+
size 280039274
|
data/fold2-00000-of-00001.parquet
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:a779daf86cd3ebd25e885e50ec131b7d05e53ad3a6ada21e387d4bc2f9d2b3d8
|
| 3 |
+
size 264174099
|
data/fold3-00000-of-00001.parquet
ADDED
|
@@ -0,0 +1,3 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
version https://git-lfs.github.com/spec/v1
|
| 2 |
+
oid sha256:5684f09517e81ff18e570608a54741e4e6715a93cbe08e32dbec3d60513457a0
|
| 3 |
+
size 289256878
|
gen_script.py
ADDED
|
@@ -0,0 +1,127 @@
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| 1 |
+
from collections.abc import Generator
|
| 2 |
+
from pathlib import Path
|
| 3 |
+
from typing import Any
|
| 4 |
+
|
| 5 |
+
import datasets
|
| 6 |
+
import numpy as np
|
| 7 |
+
from datasets import Dataset
|
| 8 |
+
from datasets.splits import NamedSplit
|
| 9 |
+
from numpy.typing import NDArray
|
| 10 |
+
from PIL import Image
|
| 11 |
+
from tqdm import tqdm
|
| 12 |
+
|
| 13 |
+
|
| 14 |
+
tissue_map = {
|
| 15 |
+
"Bile-duct": "Bile Duct",
|
| 16 |
+
"HeadNeck": "Head & Neck",
|
| 17 |
+
"Adrenal_gland": "Adrenal Gland",
|
| 18 |
+
}
|
| 19 |
+
|
| 20 |
+
features = datasets.Features(
|
| 21 |
+
{
|
| 22 |
+
"image": datasets.Image(mode="RGB"),
|
| 23 |
+
"instances": datasets.Sequence(datasets.Image(mode="1")),
|
| 24 |
+
"categories": datasets.Sequence(
|
| 25 |
+
datasets.ClassLabel(
|
| 26 |
+
num_classes=5,
|
| 27 |
+
names=[
|
| 28 |
+
"Neoplastic",
|
| 29 |
+
"Inflammatory",
|
| 30 |
+
"Connective",
|
| 31 |
+
"Dead",
|
| 32 |
+
"Epithelial",
|
| 33 |
+
],
|
| 34 |
+
)
|
| 35 |
+
),
|
| 36 |
+
"tissue": datasets.ClassLabel(
|
| 37 |
+
num_classes=19,
|
| 38 |
+
names=[
|
| 39 |
+
"Adrenal Gland",
|
| 40 |
+
"Bile Duct",
|
| 41 |
+
"Bladder",
|
| 42 |
+
"Breast",
|
| 43 |
+
"Cervix",
|
| 44 |
+
"Colon",
|
| 45 |
+
"Esophagus",
|
| 46 |
+
"Head & Neck",
|
| 47 |
+
"Kidney",
|
| 48 |
+
"Liver",
|
| 49 |
+
"Lung",
|
| 50 |
+
"Ovarian",
|
| 51 |
+
"Pancreatic",
|
| 52 |
+
"Prostate",
|
| 53 |
+
"Skin",
|
| 54 |
+
"Stomach",
|
| 55 |
+
"Testis",
|
| 56 |
+
"Thyroid",
|
| 57 |
+
"Uterus",
|
| 58 |
+
],
|
| 59 |
+
),
|
| 60 |
+
}
|
| 61 |
+
)
|
| 62 |
+
|
| 63 |
+
|
| 64 |
+
def one_hot_mask(
|
| 65 |
+
mask: NDArray[np.float64],
|
| 66 |
+
) -> tuple[NDArray[np.bool], NDArray[np.uint8]]:
|
| 67 |
+
"""Converts a mask to one-hot encoding.
|
| 68 |
+
|
| 69 |
+
Returns:
|
| 70 |
+
A dictionary with the following keys:
|
| 71 |
+
- masks: A 3D array with shape (num_masks, height, width) containing the
|
| 72 |
+
one-hot encoded masks.
|
| 73 |
+
- labels: A 1D array with shape (num_masks,) containing the class labels.
|
| 74 |
+
"""
|
| 75 |
+
masks: list[NDArray[np.bool]] = []
|
| 76 |
+
labels: list[NDArray[np.uint8]] = []
|
| 77 |
+
|
| 78 |
+
for c in range(mask.shape[-1] - 1):
|
| 79 |
+
masks.append(mask[..., c] == np.unique(mask[..., c])[1:, None, None])
|
| 80 |
+
labels.append(np.full(masks[-1].shape[0], c, dtype=np.uint8))
|
| 81 |
+
|
| 82 |
+
return np.concatenate(masks), np.concatenate(labels)
|
| 83 |
+
|
| 84 |
+
|
| 85 |
+
def process(path: str, subfolder: str) -> Generator[dict[str, Any], None, None]:
|
| 86 |
+
images = np.load(Path(path, "images", subfolder, "images.npy"), mmap_mode="r")
|
| 87 |
+
masks = np.load(Path(path, "masks", subfolder, "masks.npy"), mmap_mode="r")
|
| 88 |
+
types = np.load(Path(path, "images", subfolder, "types.npy"))
|
| 89 |
+
|
| 90 |
+
for image, mask, tissue in tqdm(
|
| 91 |
+
zip(images, masks, types, strict=True), total=len(images)
|
| 92 |
+
):
|
| 93 |
+
mask, labels = one_hot_mask(mask)
|
| 94 |
+
|
| 95 |
+
yield {
|
| 96 |
+
"image": Image.fromarray(image.astype(np.uint8)),
|
| 97 |
+
"instances": [Image.fromarray(m) for m in mask],
|
| 98 |
+
"categories": labels,
|
| 99 |
+
"tissue": tissue_map.get(tissue, tissue),
|
| 100 |
+
}
|
| 101 |
+
|
| 102 |
+
|
| 103 |
+
if __name__ == "__main__":
|
| 104 |
+
fold1 = Dataset.from_generator(
|
| 105 |
+
process,
|
| 106 |
+
gen_kwargs={"path": "PanNuke/Fold 1", "subfolder": "fold1"},
|
| 107 |
+
features=features,
|
| 108 |
+
split=NamedSplit("fold1"),
|
| 109 |
+
keep_in_memory=True,
|
| 110 |
+
)
|
| 111 |
+
fold1.push_to_hub("RationAI/PanNuke")
|
| 112 |
+
fold2 = Dataset.from_generator(
|
| 113 |
+
process,
|
| 114 |
+
gen_kwargs={"path": "PanNuke/Fold 2", "subfolder": "fold2"},
|
| 115 |
+
features=features,
|
| 116 |
+
split=NamedSplit("fold2"),
|
| 117 |
+
keep_in_memory=True,
|
| 118 |
+
)
|
| 119 |
+
fold2.push_to_hub("RationAI/PanNuke")
|
| 120 |
+
fold3 = Dataset.from_generator(
|
| 121 |
+
process,
|
| 122 |
+
gen_kwargs={"path": "PanNuke/Fold 3", "subfolder": "fold3"},
|
| 123 |
+
features=features,
|
| 124 |
+
split=NamedSplit("fold3"),
|
| 125 |
+
keep_in_memory=True,
|
| 126 |
+
)
|
| 127 |
+
fold3.push_to_hub("RationAI/PanNuke")
|