|
--- |
|
tags: |
|
- monai |
|
- medical |
|
library_name: monai |
|
license: apache-2.0 |
|
--- |
|
# Model Overview |
|
A pre-trained model for classifying nuclei cells as the following types |
|
- Other |
|
- Inflammatory |
|
- Epithelial |
|
- Spindle-Shaped |
|
|
|
This model is trained using [DenseNet121](https://docs.monai.io/en/latest/networks.html#densenet121) over [ConSeP](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet) dataset. |
|
|
|
## Data |
|
The training dataset is from https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet |
|
```commandline |
|
wget https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip |
|
unzip -q consep_dataset.zip |
|
``` |
|
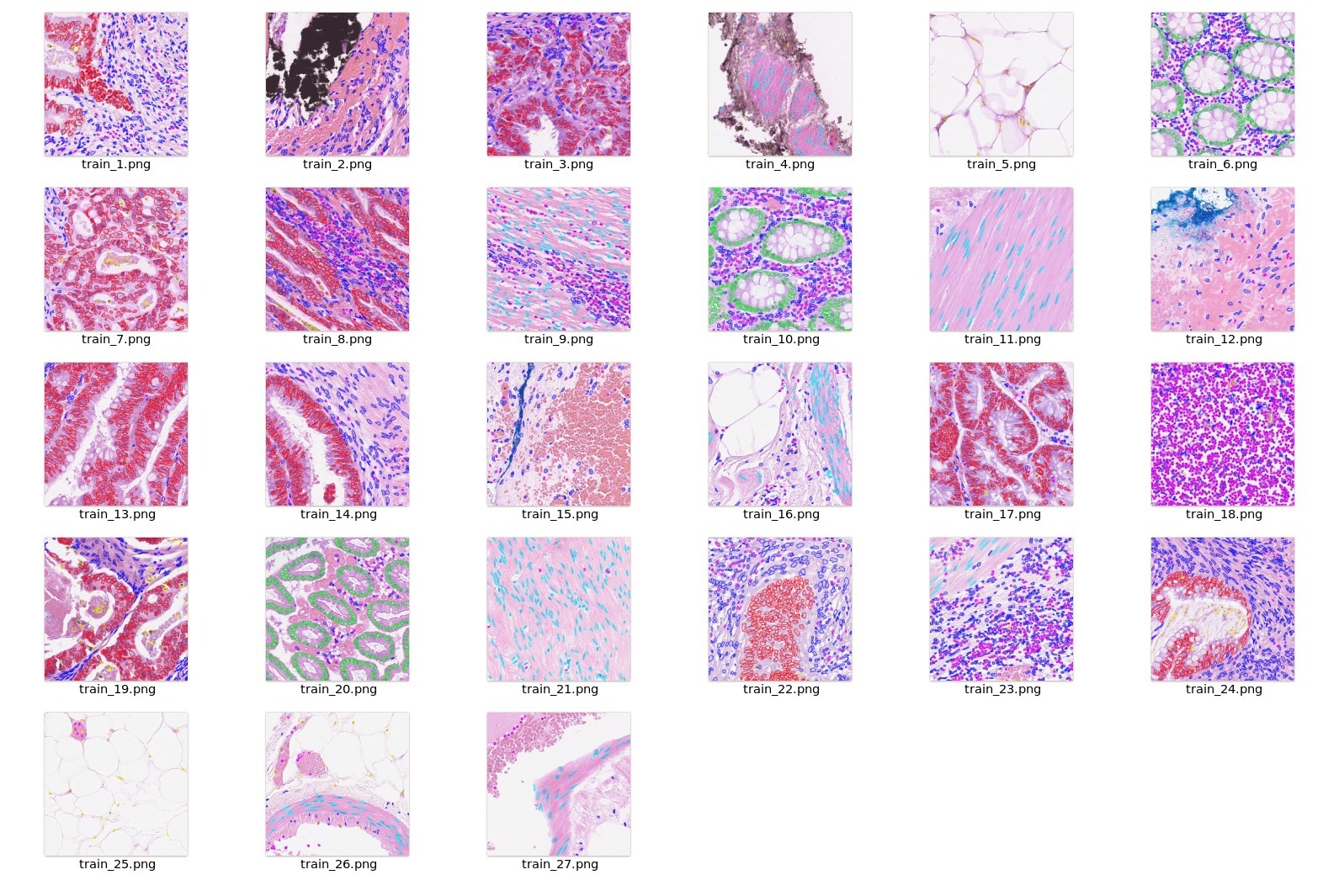<br/> |
|
|
|
### Preprocessing |
|
After [downloading this dataset](https://warwick.ac.uk/fac/cross_fac/tia/data/hovernet/consep_dataset.zip), |
|
python script `data_process.py` from `scripts` folder can be used to preprocess and generate the final dataset for training. |
|
|
|
```commandline |
|
python scripts/data_process.py --input /path/to/data/CoNSeP --output /path/to/data/CoNSePNuclei |
|
``` |
|
|
|
After generating the output files, please modify the `dataset_dir` parameter specified in `configs/train.json` and `configs/inference.json` to reflect the output folder which contains new dataset.json. |
|
|
|
Class values in dataset are |
|
|
|
- 1 = other |
|
- 2 = inflammatory |
|
- 3 = healthy epithelial |
|
- 4 = dysplastic/malignant epithelial |
|
- 5 = fibroblast |
|
- 6 = muscle |
|
- 7 = endothelial |
|
|
|
As part of pre-processing, the following steps are executed. |
|
|
|
- Crop and Extract each nuclei Image + Label (128x128) based on the centroid given in the dataset. |
|
- Combine classes 3 & 4 into the epithelial class and 5,6 & 7 into the spindle-shaped class. |
|
- Update the label index for the target nuclie based on the class value |
|
- Other cells which are part of the patch are modified to have label idex = 255 |
|
|
|
Example `dataset.json` in output folder: |
|
```json |
|
{ |
|
"training": [ |
|
{ |
|
"image": "/workspace/data/CoNSePNuclei/Train/Images/train_1_3_0001.png", |
|
"label": "/workspace/data/CoNSePNuclei/Train/Labels/train_1_3_0001.png", |
|
"nuclei_id": 1, |
|
"mask_value": 3, |
|
"centroid": [ |
|
64, |
|
64 |
|
] |
|
} |
|
], |
|
"validation": [ |
|
{ |
|
"image": "/workspace/data/CoNSePNuclei/Test/Images/test_1_3_0001.png", |
|
"label": "/workspace/data/CoNSePNuclei/Test/Labels/test_1_3_0001.png", |
|
"nuclei_id": 1, |
|
"mask_value": 3, |
|
"centroid": [ |
|
64, |
|
64 |
|
] |
|
} |
|
] |
|
} |
|
``` |
|
|
|
## Training configuration |
|
The training was performed with the following: |
|
|
|
- GPU: at least 12GB of GPU memory |
|
- Actual Model Input: 4 x 128 x 128 |
|
- AMP: True |
|
- Optimizer: Adam |
|
- Learning Rate: 1e-4 |
|
- Loss: torch.nn.CrossEntropyLoss |
|
- Dataset Manager: CacheDataset |
|
|
|
### Memory Consumption Warning |
|
|
|
If you face memory issues with CacheDataset, you can either switch to a regular Dataset class or lower the caching rate `cache_rate` in the configurations within range [0, 1] to minimize the System RAM requirements. |
|
|
|
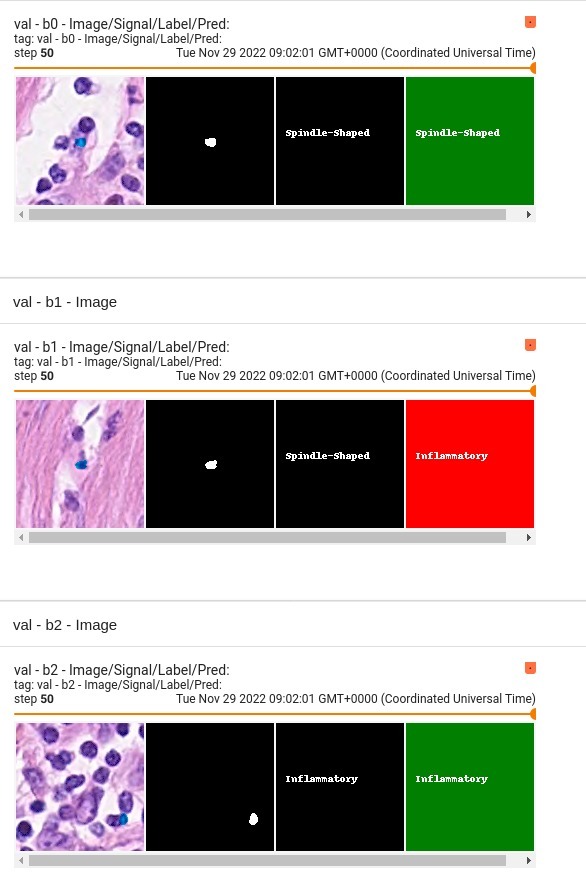
## Input |
|
4 channels |
|
- 3 RGB channels |
|
- 1 signal channel (label mask) |
|
|
|
## Output |
|
4 channels |
|
- 0 = Other |
|
- 1 = Inflammatory |
|
- 2 = Epithelial |
|
- 3 = Spindle-Shaped |
|
|
|
 |
|
|
|
## Performance |
|
This model achieves the following F1 score on the validation data provided as part of the dataset: |
|
|
|
- Train F1 score = 0.926 |
|
- Validation F1 score = 0.852 |
|
|
|
<hr/> |
|
Confusion Metrics for <b>Validation</b> for individual classes are: |
|
|
|
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped | |
|
|-----------|--------|--------------|------------|----------------| |
|
| Precision | 0.6909 | 0.7773 | 0.9078 | 0.8478 | |
|
| Recall | 0.2754 | 0.7831 | 0.9533 | 0.8514 | |
|
| F1-score | 0.3938 | 0.7802 | 0.9300 | 0.8496 | |
|
|
|
|
|
<hr/> |
|
Confusion Metrics for <b>Training</b> for individual classes are: |
|
|
|
| Metric | Other | Inflammatory | Epithelial | Spindle-Shaped | |
|
|-----------|--------|--------------|------------|----------------| |
|
| Precision | 0.8000 | 0.9076 | 0.9560 | 0.9019 | |
|
| Recall | 0.6512 | 0.9028 | 0.9690 | 0.8989 | |
|
| F1-score | 0.7179 | 0.9052 | 0.9625 | 0.9004 | |
|
|
|
|
|
|
|
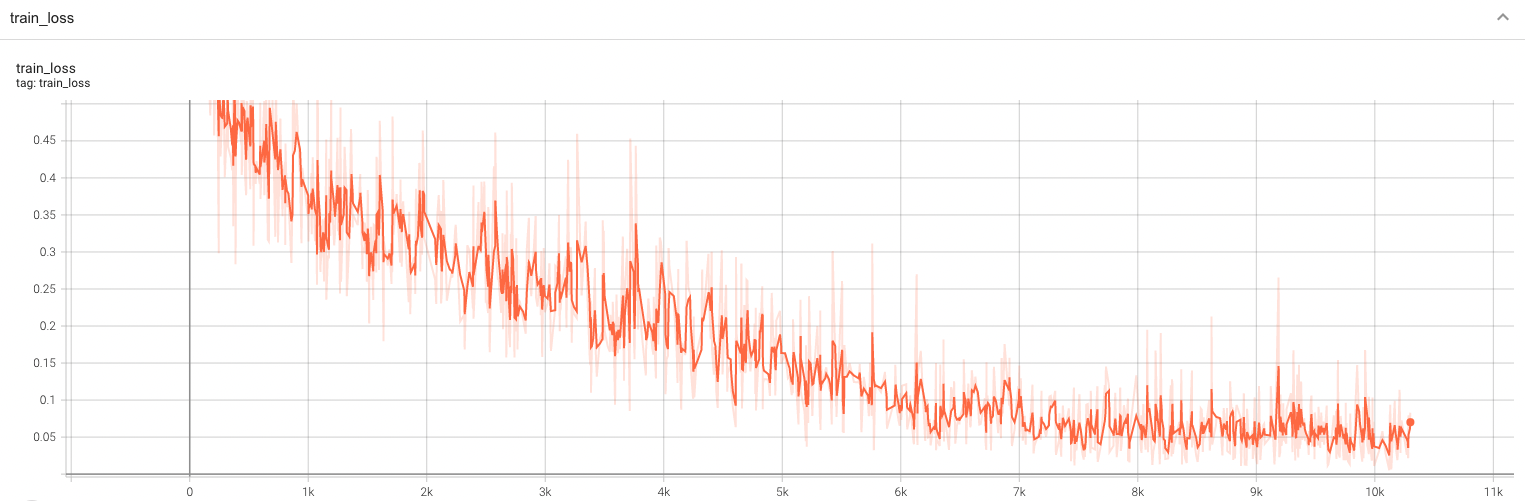
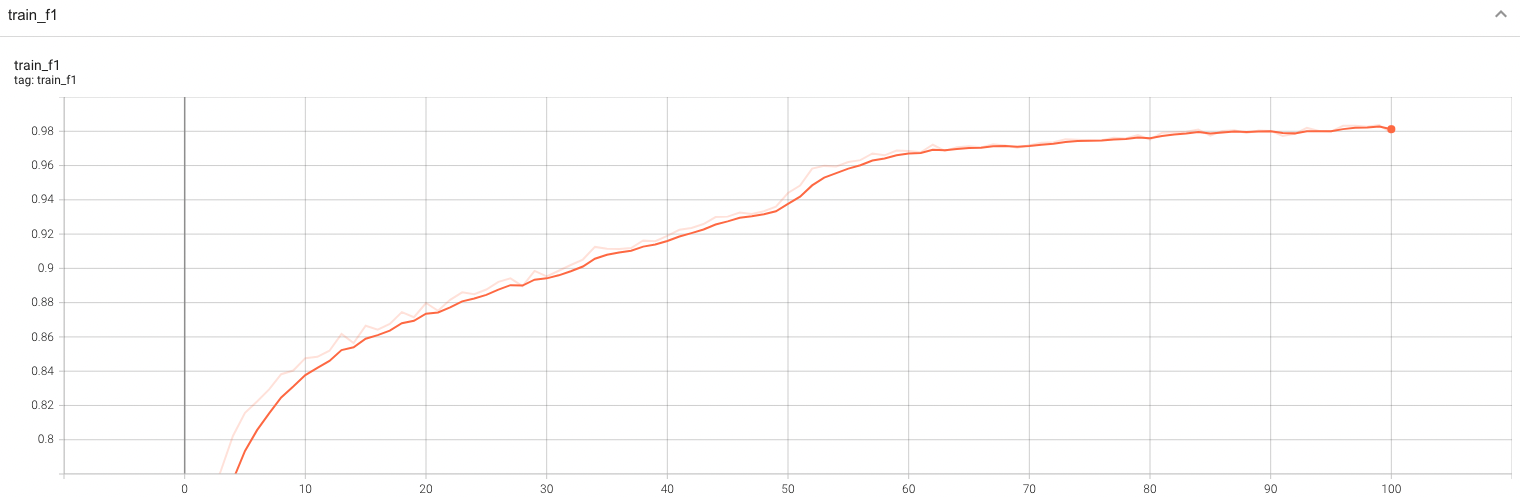
#### Training Loss and F1 |
|
A graph showing the training Loss and F1-score over 100 epochs. |
|
|
|
 <br> |
|
 <br> |
|
|
|
#### Validation F1 |
|
A graph showing the validation F1-score over 100 epochs. |
|
|
|
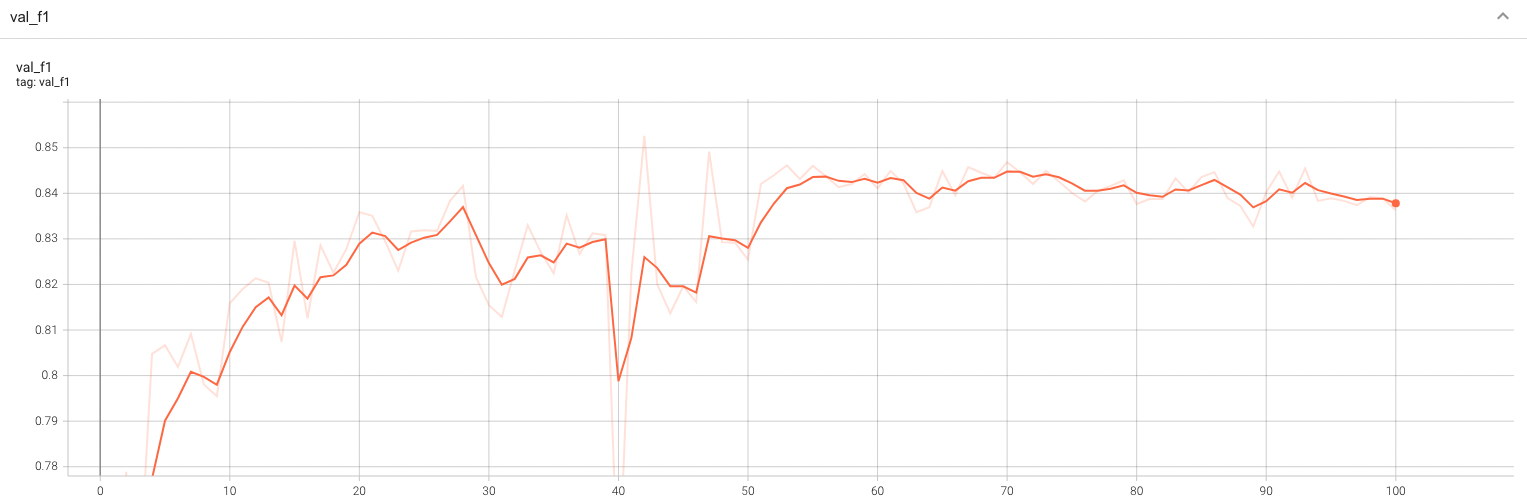 <br> |
|
|
|
## MONAI Bundle Commands |
|
In addition to the Pythonic APIs, a few command line interfaces (CLI) are provided to interact with the bundle. The CLI supports flexible use cases, such as overriding configs at runtime and predefining arguments in a file. |
|
|
|
For more details usage instructions, visit the [MONAI Bundle Configuration Page](https://docs.monai.io/en/latest/config_syntax.html). |
|
|
|
#### Execute training: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/train.json |
|
``` |
|
|
|
Please note that if the default dataset path is not modified with the actual path in the bundle config files, you can also override it by using `--dataset_dir`: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/train.json --dataset_dir <actual dataset path> |
|
``` |
|
|
|
#### Override the `train` config to execute multi-GPU training: |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/multi_gpu_train.json']" |
|
``` |
|
|
|
Please note that the distributed training-related options depend on the actual running environment; thus, users may need to remove `--standalone`, modify `--nnodes`, or do some other necessary changes according to the machine used. For more details, please refer to [pytorch's official tutorial](https://pytorch.org/tutorials/intermediate/ddp_tutorial.html). |
|
|
|
#### Override the `train` config to execute evaluation with the trained model: |
|
|
|
``` |
|
python -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json']" |
|
``` |
|
|
|
#### Override the `train` config and `evaluate` config to execute multi-GPU evaluation: |
|
|
|
``` |
|
torchrun --standalone --nnodes=1 --nproc_per_node=2 -m monai.bundle run --config_file "['configs/train.json','configs/evaluate.json','configs/multi_gpu_evaluate.json']" |
|
``` |
|
|
|
#### Execute inference: |
|
|
|
``` |
|
python -m monai.bundle run --config_file configs/inference.json |
|
``` |
|
|
|
# References |
|
[1] S. Graham, Q. D. Vu, S. E. A. Raza, A. Azam, Y-W. Tsang, J. T. Kwak and N. Rajpoot. "HoVer-Net: Simultaneous Segmentation and Classification of Nuclei in Multi-Tissue Histology Images." Medical Image Analysis, Sept. 2019. [[doi](https://doi.org/10.1016/j.media.2019.101563)] |
|
|
|
# License |
|
Copyright (c) MONAI Consortium |
|
|
|
Licensed under the Apache License, Version 2.0 (the "License"); |
|
you may not use this file except in compliance with the License. |
|
You may obtain a copy of the License at |
|
|
|
http://www.apache.org/licenses/LICENSE-2.0 |
|
|
|
Unless required by applicable law or agreed to in writing, software |
|
distributed under the License is distributed on an "AS IS" BASIS, |
|
WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. |
|
See the License for the specific language governing permissions and |
|
limitations under the License. |
|
|